Here we provide information on our VertLife data products — as they go live.
Phylogenies
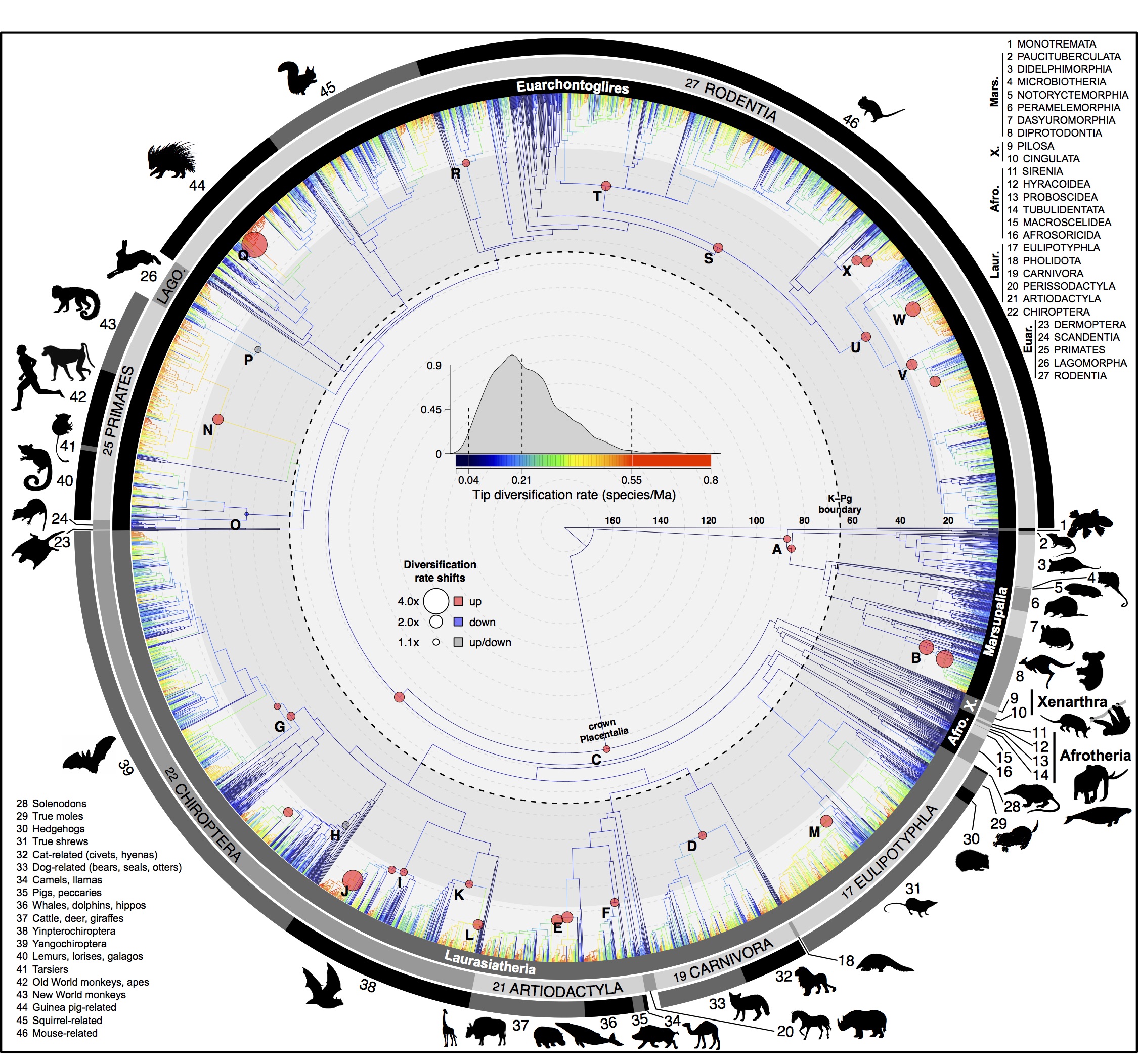
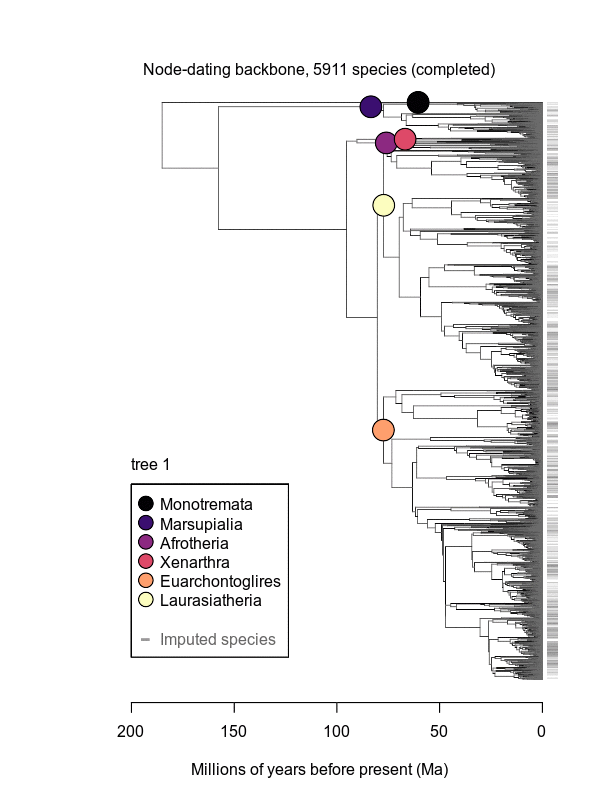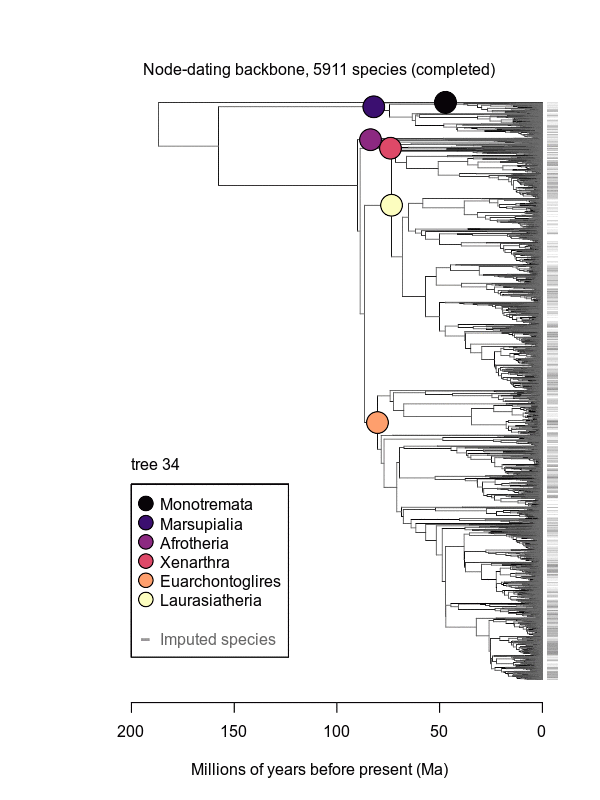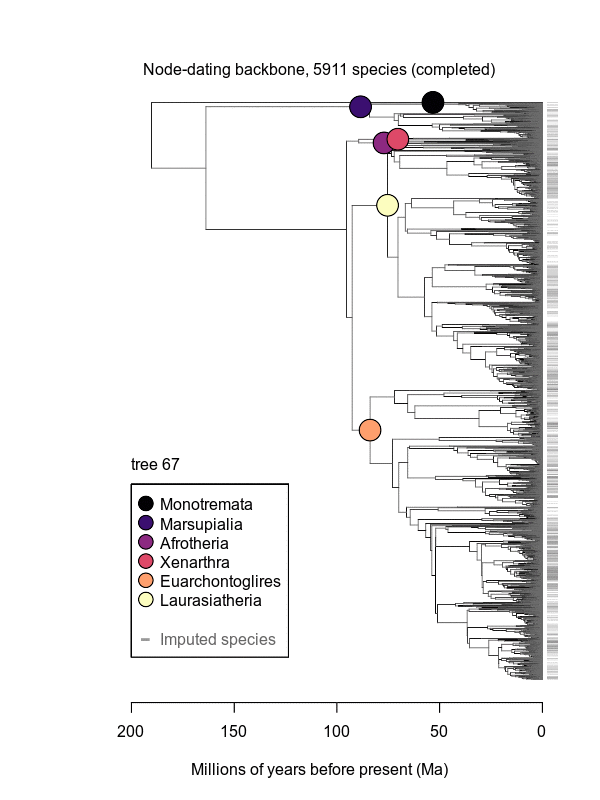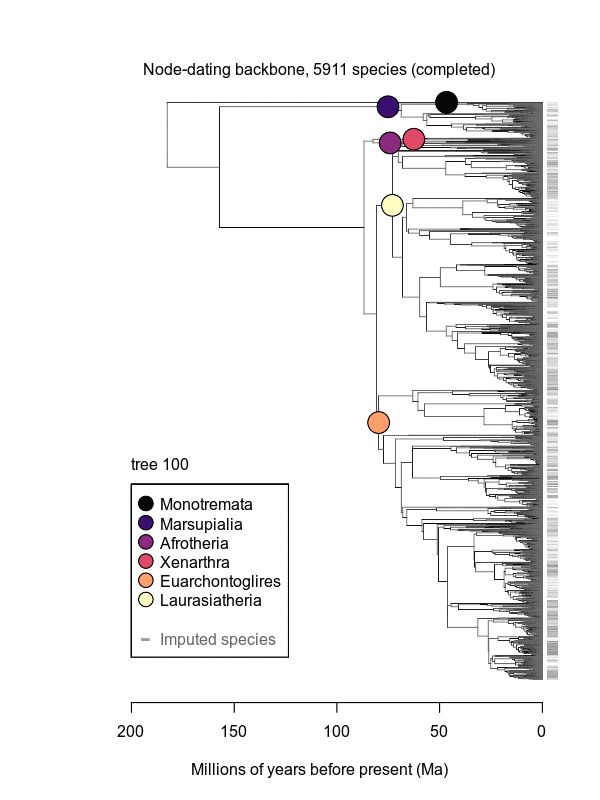
Mammals
Citation: Upham, N. S., J. A. Esselstyn, and W. Jetz. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLOS Biology. https://doi.org/10.1371/journal.pbio.3000494
Supplementary files:
- Dryad once the article is published at https://doi.org/10.5061/dryad.tb03d03.
- Taxonomic subsetting and download of mammal trees here.
- All code for running analyses will be available at https://github.com/n8upham/MamPhy_v1
Abstract: Big, time-scaled phylogenies are fundamental to connecting evolutionary processes to modern biodiversity patterns. Yet inferring reliable phylogenetic trees for thousands of species involves numerous trade-offs that have limited their utility to comparative biologists. To establish a robust evolutionary timescale for all ~6000 living species of mammals, we developed credible sets of trees that capture root-to-tip uncertainty in topology and divergence times. Our ‘backbone-and-patch’ approach to tree-building applies a newly assembled 31-gene supermatrix to two levels of Bayesian inference: (i) backbone relationships and ages among major lineages, using fossil node- or tip-dating; and (ii) species-level ‘patch’ phylogenies with non-overlapping in-groups that each correspond to one representative lineage in the backbone. Species unsampled for DNA are either excluded (‘DNA-only’ trees) or imputed within taxonomic constraints using branch lengths drawn from local birth-death models (‘completed’ trees). Joining time-scaled patches to backbones results in species-level trees of extant Mammalia with all branches estimated under the same modeling framework, thereby facilitating rate comparisons among lineages as disparate as marsupials and placentals. We compare our phylogenetic trees to previous estimates of mammal-wide phylogeny and divergence times, finding that (i) node ages are broadly concordant among studies, and (ii) recent (tip-level) rates of speciation are estimated more accurately in our study than in previous ‘supertree’ approaches where unresolved nodes led to branch length artifacts. Credible sets of mammalian phylogenetic history are now available for download at /phylosubsets, enabling investigations of long-standing questions in comparative biology.
Amphibians
Citation: Jetz, W., and R. A. Pyron. 2018. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nature Ecology & Evolution:1. https://www.nature.com/articles/s41559-018-0515-5
Supplementary files:
Abstract: Human activities continue to erode the tree of life, requiring us to prioritize research and conservation. Amphibians represent key victims and bellwethers of global change, and the need for action to conserve them is drastically outpacing knowledge. We provide a phylogeny incorporating nearly all extant amphibians (7,238 species). Current amphibian diversity is composed of both older, depauperate lineages and extensive, more recent tropical radiations found in select clades. Frog and salaman- der diversification increased strongly after the Cretaceous–Palaeogene boundary, preceded by a potential mass-extinction event in salamanders. Diversification rates of subterranean caecilians varied little over time. Biogeographically, the Afro- and Neotropics harbour a particularly high proportion of Gondwanan relicts, comprising species with high evolutionary distinctive- ness (ED). These high-ED species represent a large portion of the branches in the present tree: around 28% of all phylogenetic diversity comes from species in the top 10% of ED. The association between ED and imperilment is weak, but many species with high ED are now imperilled or lack formal threat status, suggesting opportunities for integrating evolutionary position and phylogenetic heritage in addressing the current extinction crisis. By providing a phylogenetic estimate for extant amphibians and identifying their threats and ED, we offer a preliminary basis for a quantitatively informed global approach to conserving the amphibian tree of life.
Squamates
Citation: Tonini JFR, Beard KH, Ferreira RB, Jetz W, Pyron RA (2016) Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biological Conservation, online in advance of print. http://dx.doi.org/10.1016/j.biocon.2016.03.039
Download data on Dryad here
Link to squamate phylogeny gif here
Abstract: Macroevolutionary rates of diversification and anthropogenic extinction risk differ vastly throughout the Tree of Life. This results in a highly heterogeneous distribution of Evolutionary distinctiveness (ED) and threat status among species. We examine the phylogenetic distribution of ED and threat status for squamates (amphisbaenians, lizards, and snakes) using fully-sampled phylogenies containing 9574 species and expert-based estimates of threat status for ~4000 species. We ask whether threatened species are more closely related than would be expected by chance and whether high-risk species represent a disproportionate amount of total evolutionary history. We found currently-assessed threat status to be phylogenetically clustered at broad level in Squamata, suggesting it is critical to assess extinction risks for close relatives of threatened lineages. Our findings show no association between threat status and ED, suggesting that future extinctions may not result in a disproportionate loss of evolutionary history. Lizards in degraded tropical regions (e.g., Madagascar, India, Australia, and the West Indies) seem to be at particular risk. A low number of threatened high-ED species in areas like the Amazon, Borneo, and Papua New Guinea may be due to a dearth of adequate risk assessments. It seems we have not yet reached a tipping point of extinction risk affecting a majority of species; 63% of the assessed species are not threatened and 56% are Least Concern. Nonetheless, our results show that immediate efforts should focus on geckos, iguanas, and chameleons, representing 67% of high-ED threatened species and 57% of Unassessed high-ED lineages.
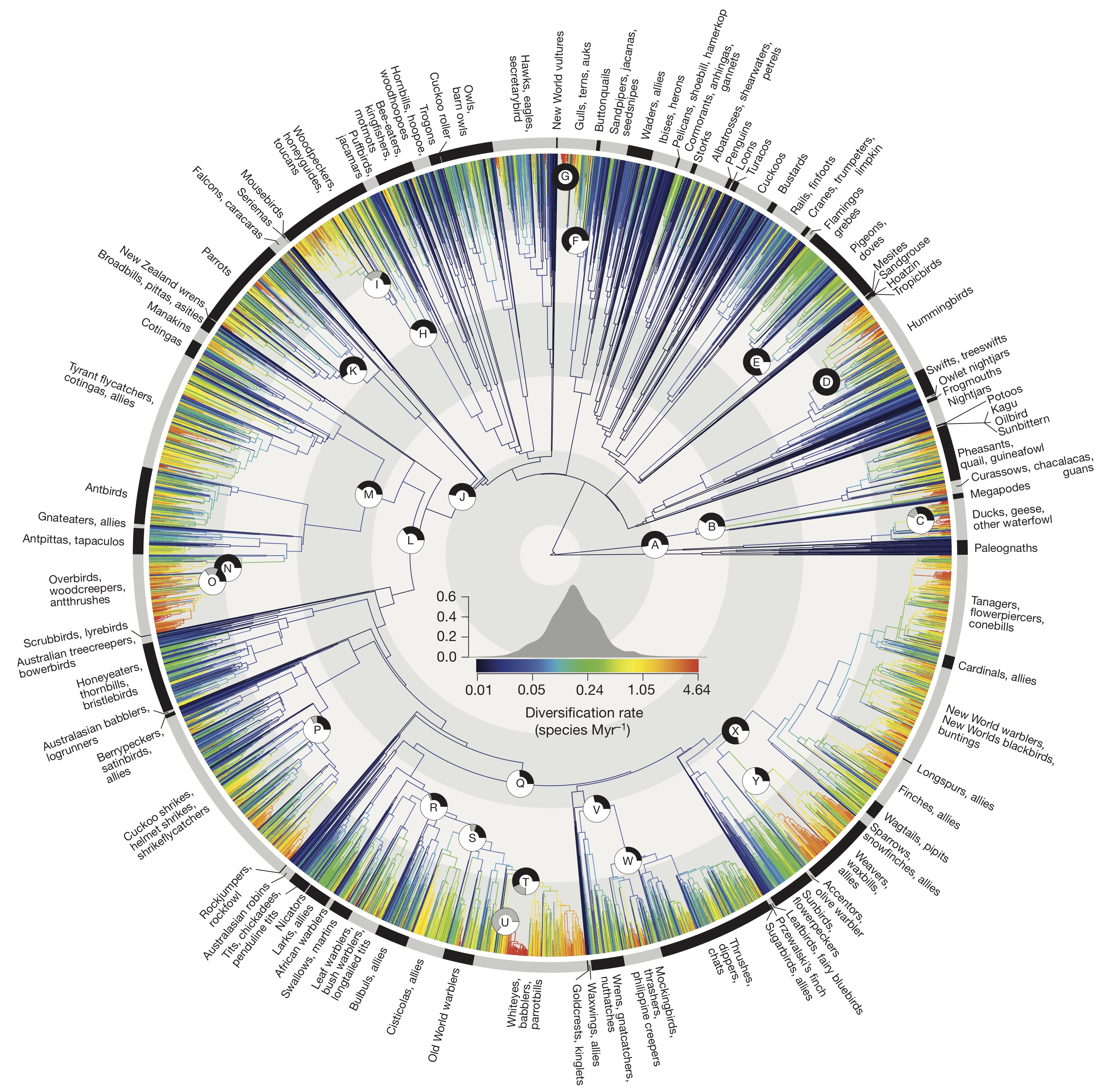
Birds
Citation: Jetz, W., G. H. Thomas, J. B. Joy, K. Hartmann, and A. O. Mooers. 2012. The global diversity of birds in space and time. Nature 491:444–448. http://www.nature.com/nature/journal/v491/n7424/abs/nature11631.html
Download data at Nature here
Abstract: Current global patterns of biodiversity result from processes that operate over both space and time and thus require an integrated macroecological and macroevolutionary perspective. Molecular time trees have advanced our understanding of the tempo and mode of diversification and have identified remarkable adaptive radiations across the tree of life. However, incomplete joint phylogenetic and geographic sampling has limited broad-scale inference. Thus, the relative prevalence of rapid radiations and the importance of their geographic settings in shaping global biodiversity patterns remain unclear. Here we present, analyse and map the first complete dated phylogeny of all 9,993 extant species of birds, a widely studied group showing many unique adaptations. We find that birds have undergone a strong increase in diversification rate from about 50 million years ago to the near present. This acceleration is due to a number of significant rate increases, both within songbirds and within other young and mostly temperate radiations including the waterfowl, gulls and woodpeckers. Importantly, species characterized with very high past diversification rates are interspersed throughout the avian tree and across geographic space. Geographically, the major differences in diversification rates are hemispheric rather than latitudinal, with bird assemblages in Asia, North America and southern South America containing a disproportionate number of species from recent rapid radiations. The contribution of rapidly radiating lineages to both temporal diversification dynamics and spatial distributions of species diversity illustrates the benefits of an inclusive geographical and taxonomical perspective. Overall, whereas constituent clades may exhibit slowdowns, the adaptive zone into which modern birds have diversified since the Cretaceous may still offer opportunities for diversification.
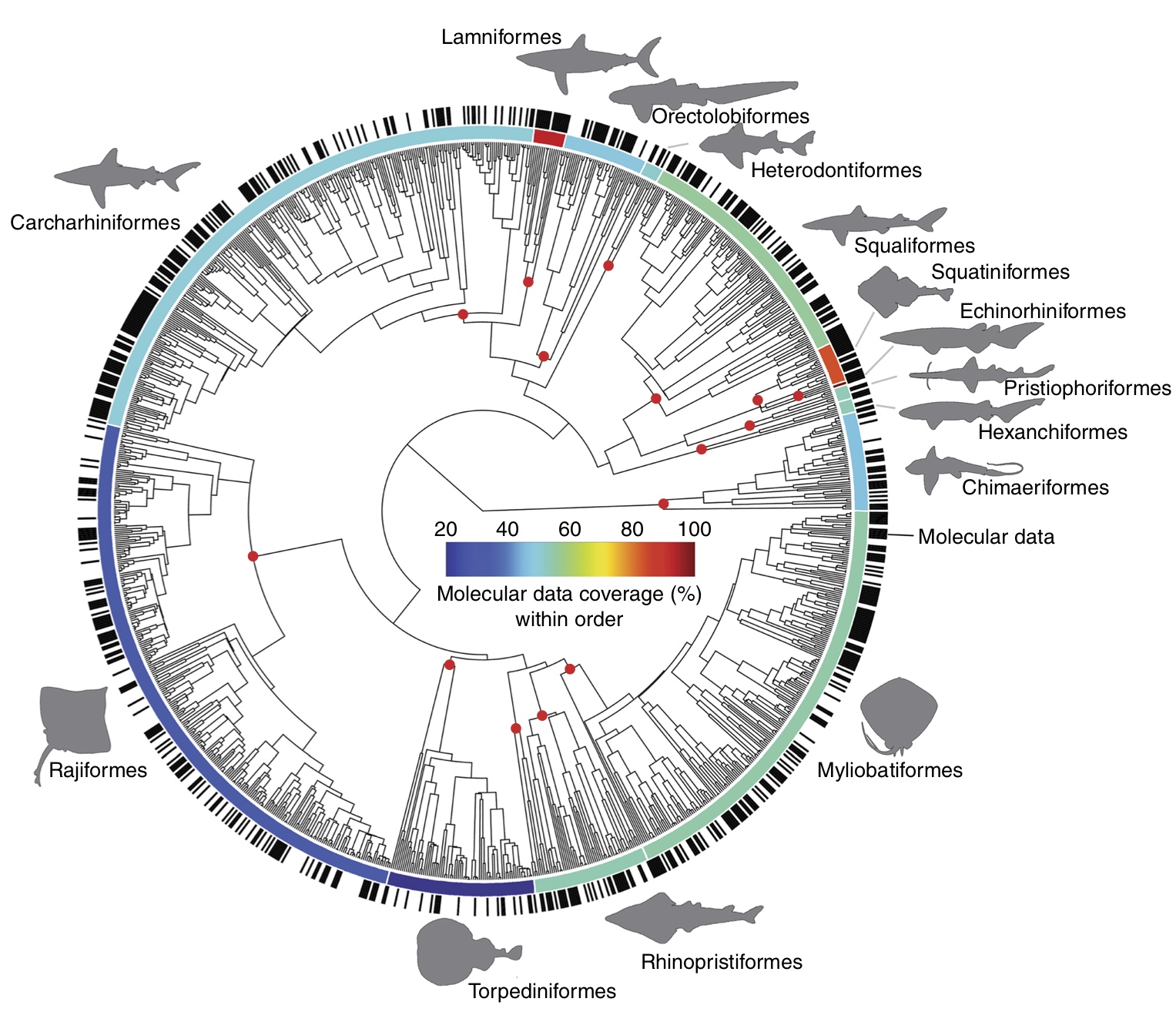
Sharks
Citation: Stein RW, Mull CG, Kuhn TS, Aschliman NC, Davidson LNK, Joy JB, Smith GJ, Dulvy NK, and Mooers AO. Global priorities for conserving the evolutionary history of sharks, rays and chimaeras. Nat Ecol Evol. 2018;2: 288–298. http://dx.doi.org/10.1038/s41559-017-0448-4
Download data:
Abstract: In an era of accelerated biodiversity loss and limited conservation resources, systematic prioritization of species and places is essential. In terrestrial vertebrates, evolutionary distinctness has been used to identify species and locations that embody the greatest share of evolutionary history. We estimate evolutionary distinctness for a large marine vertebrate radiation on a dated taxon-complete tree for all 1,192 chondrichthyan fishes (sharks, rays and chimaeras) by augmenting a new 610-species molecular phylogeny using taxonomic constraints. Chondrichthyans are by far the most evolutionarily distinct of all major radiations of jawed vertebrates—the average species embodies 26 million years of unique evolutionary history. With this metric, we identify 21 countries with the highest richness, endemism and evolutionary distinctness of threatened species as targets for conservation prioritization. On average, threatened chondrichthyans are more evolutionarily distinct—further motivating improved conservation, fisheries management and trade regulation to avoid significant pruning of the chondrichthyan tree of life.
Taxonomies
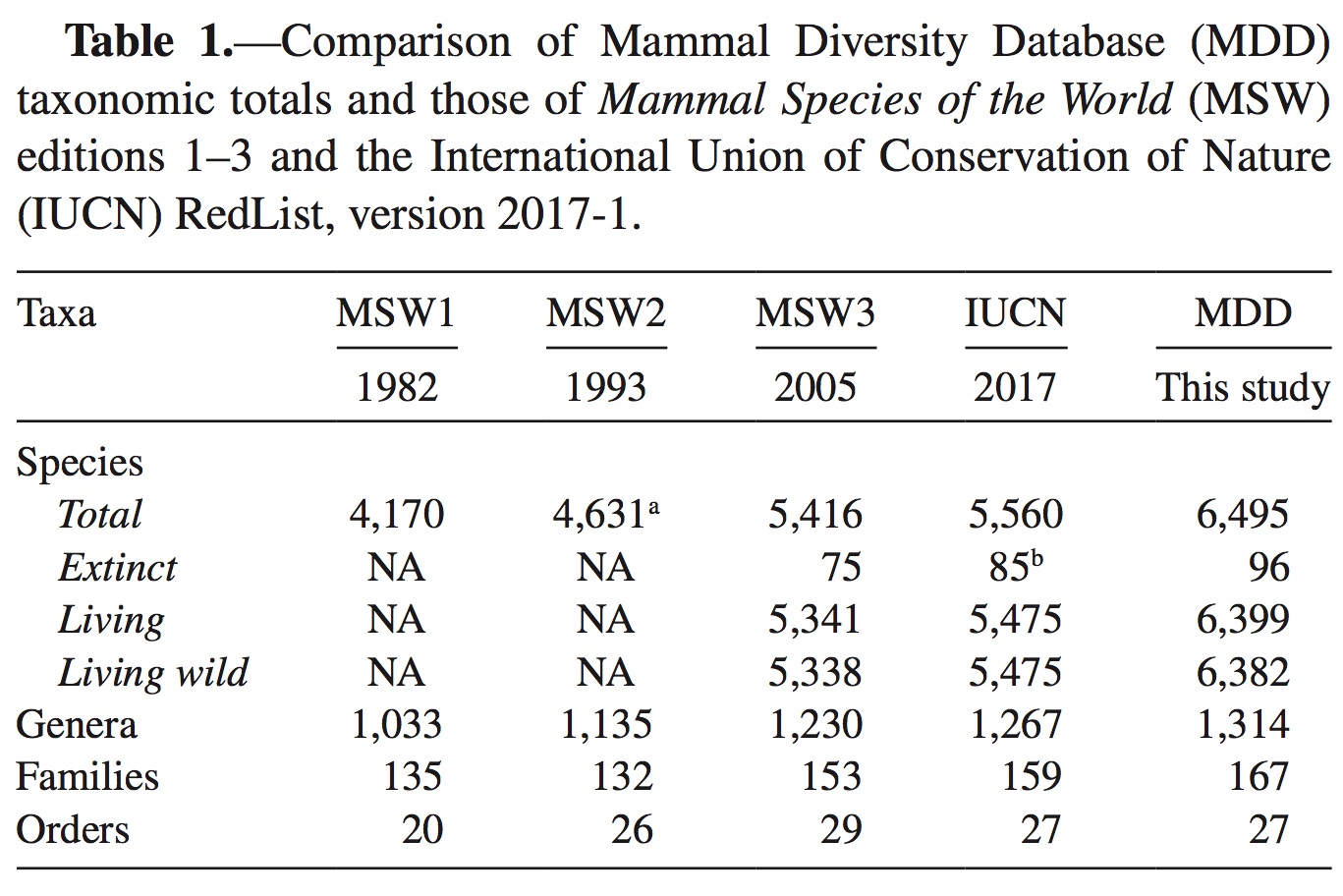
Mammals: The Mammal Diversity Database
Citation: Burgin, C. J., J. P. Colella, P. L. Kahn, and N. S. Upham. 2018. How many species of mammals are there? Journal of Mammalogy 99:1–14. https://academic.oup.com/jmammal/article/99/1/1/4834091
Logistical and planning support for this work came from the NSF Vertlife Terrestrial grant with development commissioned by the American Society of Mammalogists.
The ASM Biodiversity Committee stewards the Mammal Diversity Database, an updatable and online database of mammal taxonomic and biodiversity information. This database aims to serve the global scientific community by providing the latest information on species-level and higher taxonomic changes, thereby promoting more rigorous study of mammalian biodiversity worldwide. The initial objective for this online database is to aggregate, curate, and compile new citations on species descriptions and taxonomic revisions into regular releases that are downloadable in comma-delimited format. Downstream goals include expanded hosting of ecological, trait, and taxonomic data, and an online forum for discussing mammalian taxonomy and systematics.
Species Rates
We are providing tip rates associated with each of the VertLife phylogenetic frameworks. These include:
- Fair Proportion metric for assessing Evolutionary Distinctness, often abbreviated as ‘FP’ or ‘ED’ (Redding et al 2010)
- Equal Splits metric, often abbreviated as ‘ES’ (Redding et al 2006)
- The Species Diversification Rates metric, often abbreviated as DR or Tip DR, measuring speciation rates (Jetz et al 2012).
Tip rates are tip level aggregates of trees’ interior branch structure and lengths. For each group, we ran calculations across 1,000 trees selected at random from the provided pseudo-posterior distributions and calculated a range of measures capturing averages and among-tree variation. These include Median, Harmonic Mean, Geometric Mean, Arithmetic Mean, Range, Variance, Standard Deviation, Coefficient of Variance, and lower and upper 95 percentiles. We plan to make these metrics available through the tree subset tools – see there for updates. Please make sure to cite the respective original tree references when using these data.
Download data:
- Birds (Ericson): DR, ES, FP
- Birds (Hackett): DR, ES, FP
- Mammals: DR, ES, FP
- Amphibians: DR, ES, FP
- Squamates: DR, ES, FP
- Sharks: DR, ES, FP
References:
-
Redding, D. W. & Mooers, A. O. (2006): Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678.
-
Redding, D.W., DeWolff, C.V., and Mooers, A.Ø. (2010). Evolutionary distinctiveness, threat status, and ecological oddity in primates. Conserv. Biol. 24, 1052–1058.
-
Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO (2012). The global diversity of birds in space and time. Nature 491(7424):444-8.
Traits
TetrapodTraits with phylogeny-based multiple imputation
Citation: Moura, M.R., Ceron, K., Guedes, J.J.M., Chen-Zhao, R., Sica, Y.V., Hart, J., Dorman, W., Gonzalez-del-Pliego, P., Ranipeta, A., Catenazzi, A.., Werneck, F.P., Toledo, L.F., Upham, N.S., Tonini, J.F.R., Colston, T.J., Guralnick, R., Bowie, R.C.K., Pyron, R.A., Jetz, W. A phylogeny-informed characterisation of global tetrapod traits addresses data gaps and biases. BioRXiv, 2024, doi: 10.1101/2023.03.04.531098v3
Supplementary Files:
- Data download on Zenodo: https://zenodo.org/records/11303604
- R code for data analysis: https://doi.org/10.5281/zenodo.10582070
- Paper: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3002658
Abstract:
Tetrapods (amphibians, reptiles, birds, and mammals) are model systems for global biodiversity science, but continuing data gaps, limited data standardisation, and ongoing flux in taxonomic nomenclature constrain integrative research on this group and potentially cause biased inference. We combined and harmonised taxonomic, spatial, phylogenetic, and attribute data with phylogeny-based multiple imputation to provide a comprehensive data resource (TetrapodTraits 1.0.0) that includes values, predictions, and sources for body size, activity time, micro- and macrohabitat, ecosystem, threat status, biogeography, insularity, environmental preferences, and human influence, for all 33,281 tetrapod species covered in recent fully sampled phylogenies. We assess gaps and biases across taxa and space, finding that shared data missing in attribute values increased with taxon-level completeness and richness across clades. Prediction of missing attribute values using multiple imputation revealed substantial changes in estimated macroecological patterns. These results highlight biases incurred by nonrandom missingness and strategies to best address them. While there is an obvious need for further data collection and updates, our phylogeny-informed database of tetrapod traits can support a more comprehensive representation of tetrapod species and their attributes in ecology, evolution, and conservation research.
EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals
Citation: Hamish Wilman, Jonathan Belmaker, Jennifer Simpson, Carolina de la Rosa, Marcelo M. Rivadeneira, and Walter Jetz. 2014. EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals. Ecology 95:2027. http://dx.doi.org/10.1890/13-1917.1
Abstract: Species are characterized by physiological, behavioral, and ecological attributes that are all subject to varying evolutionary and ecological constraints and jointly determine their role and function in ecosystems. Attributes such as diet, foraging strata, foraging time, and body size, in particular, determine a large portion of the “Eltonian” niches of species. Here we present a global species-level compilation of these key attributes for all 9993 and 5400 extant bird and mammal species derived from key literature sources. Global handbooks and monographs allowed the consistent sourcing of attributes for most species. For diet and foraging stratum we followed a defined protocol to translate the verbal descriptions into standardized, semiquantitative information about relative importance of different categories. Together with body size (continuous) and activity time (categorical) this enables a much finer distinction of species’ foraging ecology than typical categorical guild assignments allow. Attributes lacking information for specific species were flagged, and interpolated values based on taxonomy were provided instead. The presented data set is limited by, among others, these select cases missing observed data, by errors and uncertainty in the expert assessment as presented in the literature, and by the lack of intraspecific information. However, the standardized and transparent nature and complete global coverage of the data set should support an array of potential studies in biogeography, community ecology, macroevolution, global change biology, and conservation. Potential uses include comparative work involving these traits as focal or secondary variables, ecological research on the trait or trophic structure of communities, or conservation science concerned with the loss of function among species or in ecosystems in a changing world. We hope that this publication will spur the sharing, collaborative curation, and extension of data to the benefit of a more integrative, rigorous, and global biodiversity science.
Discovery Potential
View the new interactive map!
View more information
Shortfalls and opportunities in terrestrial vertebrate species discovery
Citation: Moura, M.R. & W. Jetz, 2021. https://dx.doi.org/10.1038/s41559-021-01411-5
Abstract: Much of biodiversity remains undiscovered, causing species and their functions to remain unrealized and potentially lost in ignorance. Here we use extensive species-level data in a time-to-event model framework to identify taxonomic and geographic discovery gaps in terrestrial vertebrates. Biological, environmental and sociological factors all affect discovery probability and together provide strong predictive ability for species discovery. Our model identifies distinct taxonomic and geographic unevenness in future discovery potential, with greatest opportunities for amphibians and reptiles, and for Neotropical and Indo-Malayan forests. Brazil, Indonesia, Madagascar and Colombia emerge as holding greatest discovery opportunities, with a quarter of potential discoveries estimated. These findings highlight the importance of international policy support for basic taxonomic research and the potential of quantitative models to aid species discovery.
Citations
Mammals:
- Upham, N. S., J. A. Esselstyn, and W. Jetz. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLOS Biology. https://doi.org/10.1371/journal.pbio.3000494
- Upham, N. S., J. A. Esselstyn, and W. Jetz. 2019. Ecological causes of speciation and species richness in the mammal tree of life. bioRxiv:504803. https://doi.org/10.1101/504803v3
Birds:
- Jetz, W., G. H. Thomas, J. B. Joy, K. Hartmann, and A. O. Mooers. 2012. The global diversity of birds in space and time. Nature 491:444–448. http://www.nature.com/nature/journal/v491/n7424/abs/nature11631.html
Amphibians:
- Jetz, W., and R. A. Pyron. 2018. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nature Ecology & Evolution:1. https://www.nature.com/articles/s41559-018-0515-5
Squamates:
- Tonini JFR, Beard KH, Ferreira RB, Jetz W, Pyron RA (2016) Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biological Conservation, online in advance of print. http://dx.doi.org/10.1016/j.biocon.2016.03.039
Sharks:
- Stein RW, Mull CG, Kuhn TS, Aschliman NC, Davidson LNK, Joy JB, Smith GJ, Dulvy NK, and Mooers AO. Global priorities for conserving the evolutionary history of sharks, rays and chimaeras. Nat Ecol Evol. 2018;2: 288–298. http://dx.doi.org/10.1038/s41559-017-0448-4


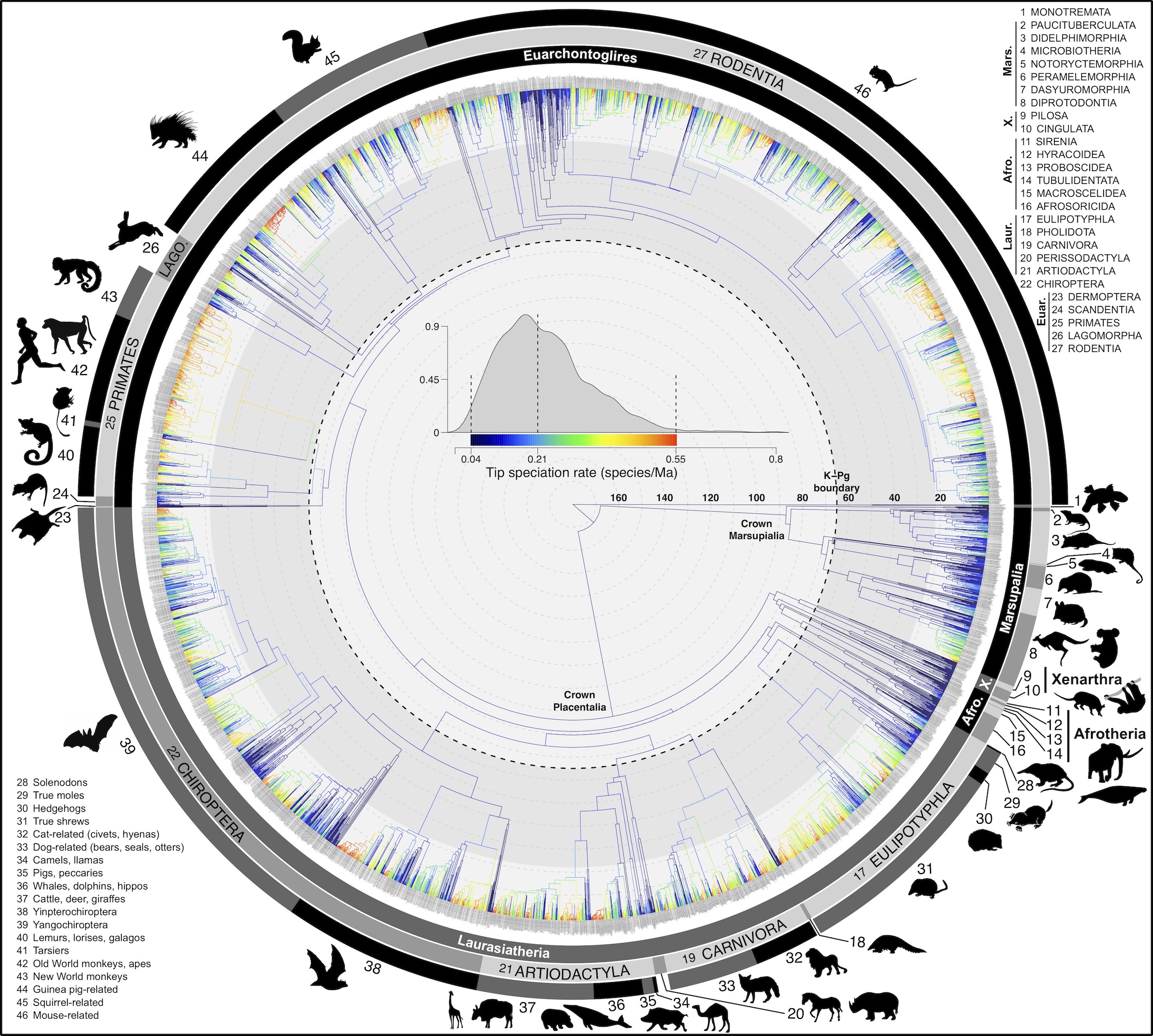





.svg.png)